This workshop will provide hands-on experience of building workflows using the Python package Crossflow and executing them on Archer2.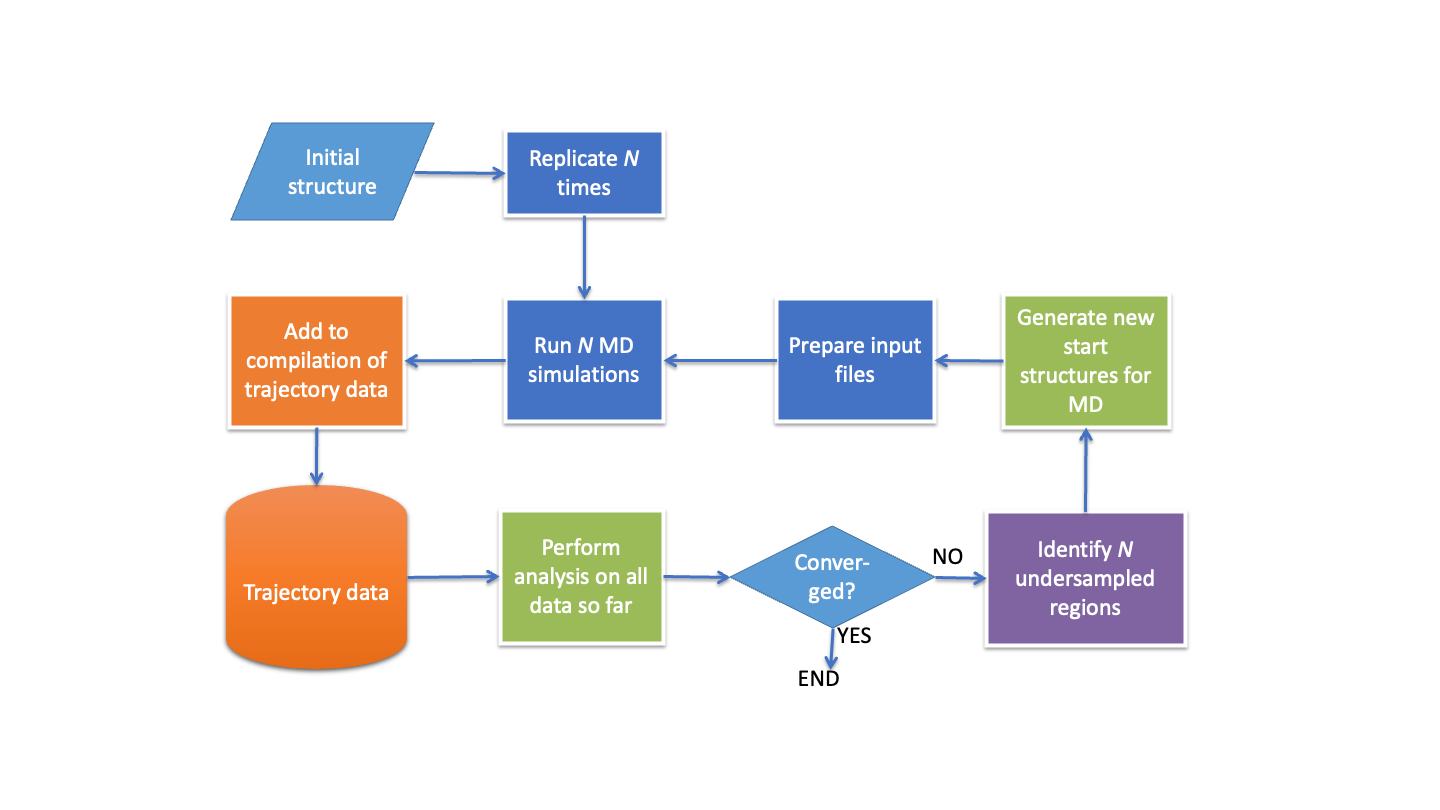
Workflows are integral to computational chemistry activity; for example, before a molecular dynamics simulation is run there are typically numerous steps involved in remediating input molecular structure data, adding solvent and/or membrane components, assigning forcefield parameters etc.; then the simulation process itself will involve a series of preliminary relaxation and equilibration steps before the production phase is reached. A variety of advanced simulation methods are of themselves iterative workflows involving cycles of data generation, analysis, and decision making; replica exchange approaches would be an example. The implementation of these workflows may be as simple as the construction of a bash script, or as complex as encoding the functionality within an existing monolithic piece of software. The first approach may enable rapid prototyping, but is seldom robust, flexible and appropriate for wide dissemination; the second can produce a high-quality product but its range of functionality is hard coded and development times are long.
Computational workflow systems are designed to cover the middle ground: to enable the relatively rapid construction of bespoke tools that offer novel functionality but which are also of a quality that makes them suitable for wide use, and use at scale. Crossflow differs from other workflow tools in that there is less reliance on providing a selection of pre-built “building blocks” for a user to assemble into the workflow they need; rather it allows users to create quickly and easily exactly the elements they require. To provide experience of this flexibility, workshop participants will learn how to build from scratch workflows for both an enhanced sampling simulation method, and for an iterative docking-based ligand optimisation approach.
Attendees will require a reasonable level of proficiency with Python, and a general knowledge of MD simulation and docking methods. Previous experience of using Archer2 or other HPC systems will be an advantage.
Venue
The workshop will be held in-person only in the Bragg building in room 1.49 at Leeds University.
Registration
Registration is £20, lunch will be provided but travel and accomodation are not covered.
Register here - deadline 1st of November.
CCPBioSim (grant no. EP/T026308/1) acknowledges the funding support it receives from EPSRC through an SLA with STFC’s Scientific Computing Department.



